M4DI
Cellular heterogeneity in biological samples is a key factor that determines disease progression, but also influences biomedical analysis of samples and patient classification. At the molecular level, the cellular composition of tissues is difficult to assess and quantify, as it is hidden within the bulk molecular profiles of samples (average profile of millions of cells), with all cells present in the tissue contributing to the recorded signal. Despite great promise, conventional computational approaches to quantifying cellular heterogeneity from mixtures of cells have encountered difficulties in providing robust and biologically relevant estimates.
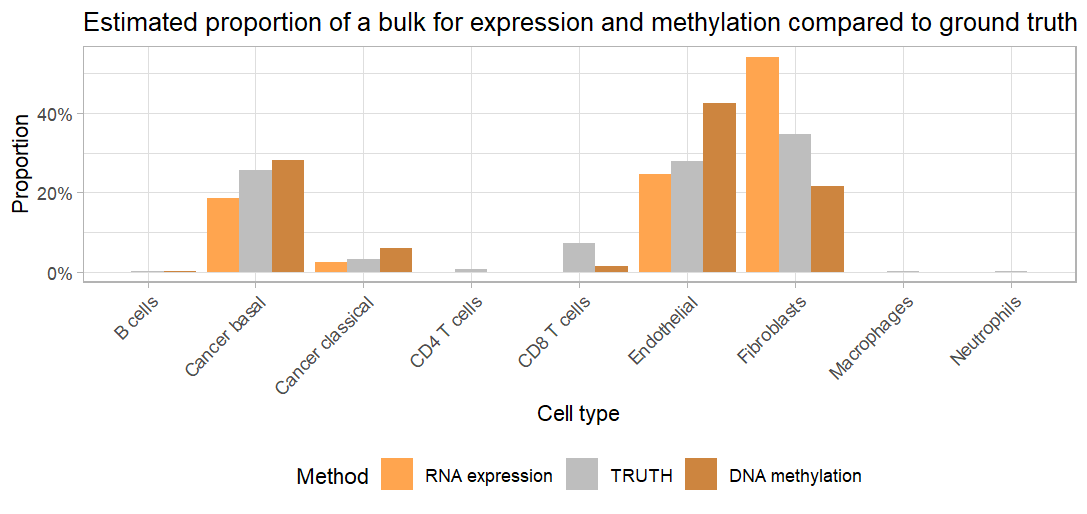
So far, most statistical methods used for cell deconvolution ignore the biological relationships between the molecular features used in the models. Our goal is to provide a statistical framework for deconvolution including (i) the stochastic dependence across molecular features induced by the mutual regulation mechanisms; (ii) the a priori knowledge of the topology of multilayer interaction networks; and (iii) the similarity between samples that may be induced by controlled experimental conditions.
So far, most statistical methods used for cell deconvolution ignore the biological relationships between the molecular features used in the models. Our goal is to provide a statistical framework for deconvolution including (i) the stochastic dependence across molecular features induced by the mutual regulation mechanisms; (ii) the a priori knowledge of the topology of multilayer interaction networks; and (iii) the similarity between samples that may be induced by controlled experimental conditions.
