CauseHet
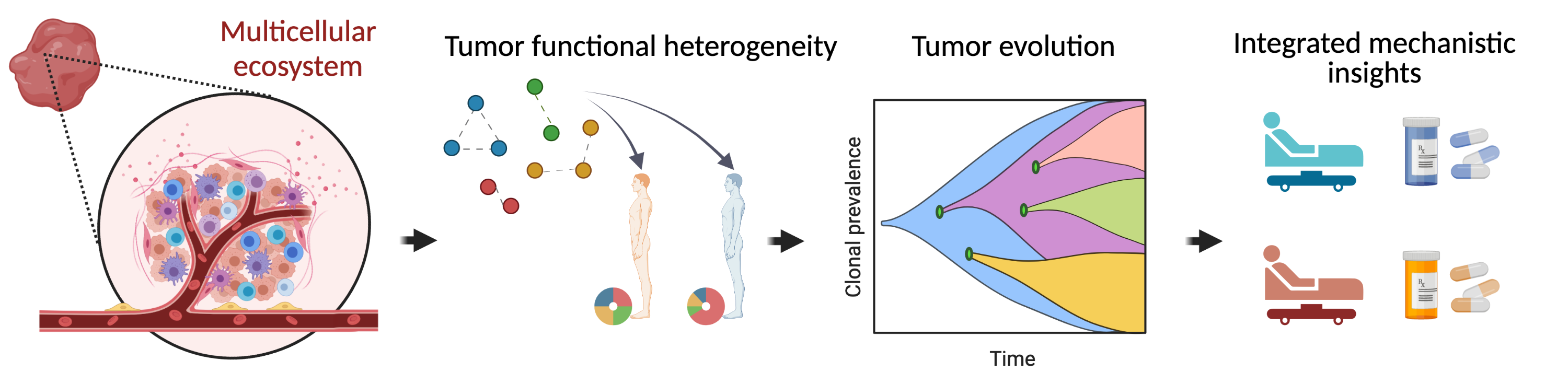
Cancers are heterogeneous diseases, each single tumor evolving as a multicellular autonomous system. Tumors are made up of cells with different identities and origins, that dynamically interact with each other to form the tumor ecosystem. Heterogeneity in tumor cellular composition is a key factor driving cancer progression but is hard to observe and quantify. So far, our restricted ability to estimate cellular heterogeneity in a given sample has hampered our understanding of its function during oncogenic processes.
Our goal is to provide a statistical framework describing tumor functional heterogeneity and analyzing its effect on tumorigenesis. Taking advantage of multiple high-throughput molecular assays combined with single-cell analysis, we will study gene deregulation at the single-tumor scale and enrich this analysis with knowledge of biological network architecture in humans. From there, we will estimate the functional heterogeneity derived from cancer and micro-environment cells. We will quantify which functional modules (i.e., groups of genes associated with biological functions) are deregulated in each tumor. In parallel, we will develop algorithms to predict the effect of functional heterogeneity on the tumor development, from both an evolutionary and a mechanistic perspective. First, we will detect associations between tumor heterogeneity and (epi)genomic alterations in cancer cells. Second, we will perform high-dimension causal inference to investigate the link between heterogeneity, patient treatment and survival. By focusing on functional heterogeneity, CauseHet has the potential to overcome challenges that have prevented the accurate quantification of intra-tumor heterogeneity. This functional vision of tumor heterogeneity will allow a better understanding of the biological mechanisms of tumorigenesis. It will offer leads to predict the impacts of tumor heterogeneity on patient response to treatment, notably in the context of personalized therapies.
Our goal is to provide a statistical framework describing tumor functional heterogeneity and analyzing its effect on tumorigenesis. Taking advantage of multiple high-throughput molecular assays combined with single-cell analysis, we will study gene deregulation at the single-tumor scale and enrich this analysis with knowledge of biological network architecture in humans. From there, we will estimate the functional heterogeneity derived from cancer and micro-environment cells. We will quantify which functional modules (i.e., groups of genes associated with biological functions) are deregulated in each tumor. In parallel, we will develop algorithms to predict the effect of functional heterogeneity on the tumor development, from both an evolutionary and a mechanistic perspective. First, we will detect associations between tumor heterogeneity and (epi)genomic alterations in cancer cells. Second, we will perform high-dimension causal inference to investigate the link between heterogeneity, patient treatment and survival. By focusing on functional heterogeneity, CauseHet has the potential to overcome challenges that have prevented the accurate quantification of intra-tumor heterogeneity. This functional vision of tumor heterogeneity will allow a better understanding of the biological mechanisms of tumorigenesis. It will offer leads to predict the impacts of tumor heterogeneity on patient response to treatment, notably in the context of personalized therapies.
