AIMPACT
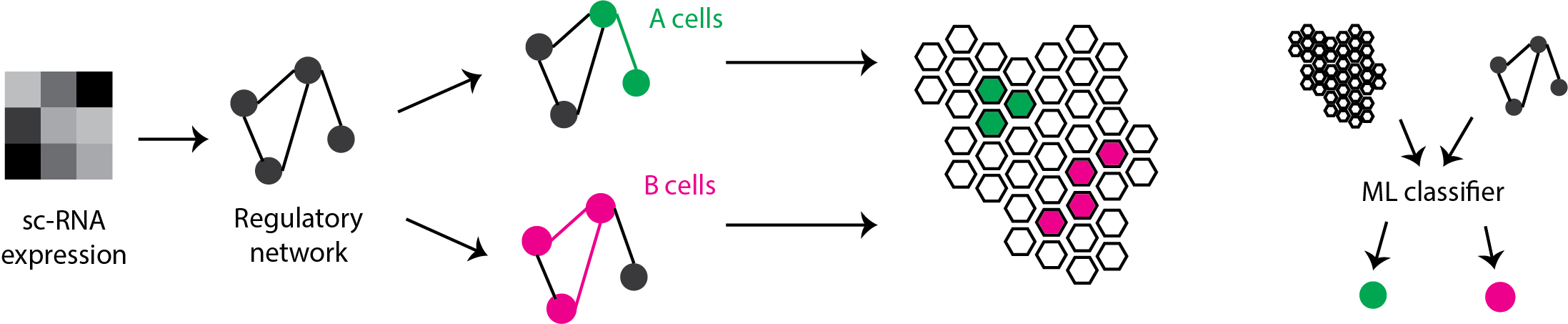
Tumor cell plasticity is the ability of cancer cells to dynamically reprogram their transcriptional programs in response to environmental cues. It is a key driver of therapy resistance and tumor evolution across multiple cancer types. The complexity of tumor cell plasticity presents a significant analytical challenge that conventional methods struggle to address due to their limited capacity to model dynamic state transitions in high-dimensional biological data. Advanced artificial intelligence approaches are uniquely positioned to decipher the complex, nonlinear relationships governing cancer cell state transitions.
Building on our established expertise in cancer biology and computational modeling, our project aims to develop breakthrough artificial intelligence methods that integrate gene regulatory network analysis, spatial transcriptomics, and explainable machine learning techniques to characterize, interpret, and predict cellular state transitions in solid tumors. We will address three key technical challenges: uncertainty quantification in biological networks, interpretability of complex AI models, and integration of spatial context with molecular dynamics. Our primary focus is on Pancreatic adenocarcinoma (PDAC), one of the most lethal malignancies, for which we have deep expertise and access to rich single-cell and spatial transcriptomic datasets. Importantly our framework is designed to be generalizable to other cancer types. Beyond methodological advances, we will establish rigorous benchmarking frameworks and develop open-source software tools to foster community adoption and scientific reproducibility. To catalyze collaborative science and accelerate innovation, we will organize scientific competitions that bring together diverse expertise from the AI and biomedical communities, creating a dynamic platform for methodological advancement and knowledge exchange. This interdisciplinary approach, at the interface between artificial intelligence, bioinformatics, and oncology, will decipher the molecular and spatial mechanisms governing tumor plasticity. Ultimately, our work will pave the way for novel therapeutic strategies and personalized medicine while contributing to the development of a national ecosystem for AI innovation in oncology research.
Building on our established expertise in cancer biology and computational modeling, our project aims to develop breakthrough artificial intelligence methods that integrate gene regulatory network analysis, spatial transcriptomics, and explainable machine learning techniques to characterize, interpret, and predict cellular state transitions in solid tumors. We will address three key technical challenges: uncertainty quantification in biological networks, interpretability of complex AI models, and integration of spatial context with molecular dynamics. Our primary focus is on Pancreatic adenocarcinoma (PDAC), one of the most lethal malignancies, for which we have deep expertise and access to rich single-cell and spatial transcriptomic datasets. Importantly our framework is designed to be generalizable to other cancer types. Beyond methodological advances, we will establish rigorous benchmarking frameworks and develop open-source software tools to foster community adoption and scientific reproducibility. To catalyze collaborative science and accelerate innovation, we will organize scientific competitions that bring together diverse expertise from the AI and biomedical communities, creating a dynamic platform for methodological advancement and knowledge exchange. This interdisciplinary approach, at the interface between artificial intelligence, bioinformatics, and oncology, will decipher the molecular and spatial mechanisms governing tumor plasticity. Ultimately, our work will pave the way for novel therapeutic strategies and personalized medicine while contributing to the development of a national ecosystem for AI innovation in oncology research.
